Telomere-to-telomere Gapless Genome Assembly
Telomere-to-telomere Gapless Genome Assembly
Blog Article
Long read long sequencing has provided strong support for genomics projects in academic circles. Several companies have driven this process, notably Pacific Biosciences (PacBio) and Oxford Nanopore Technologies (ONT). We have two state-of-the-art platforms at your disposal, PacBio SMRT Sequencing and Oxford Nanopore Sequencing. Advances in genomic technology are increasingly pushing the boundaries of our understanding of the biological world. One of these highly publicized advances is the realization of telomere-to-telomere (T2T) gapless genome assembly. This technology has revolutionized the way we look at genome research and understand the complex genetic structure of different organisms.
Telomere-to-telomere genome assembly refers to the generation of complete chromosomal DNA sequences without any gaps from one end of the chromosome (telomere) to the other. The combination of long, accurate PacBio HiFi reads and ultra-long Oxford Nanopore reads along with advances in computational algorithms has driven this technology, and achieving this level of completeness and accuracy is a huge step forward for genome research. These technologies have enabled the study of complex genomic regions that were previously difficult to address due to their highly repetitive sequences, such as mitoses and telomeres.
In March 2022, the Telomere to Telomere (T2T) Consortium announced the completion and analysis of the first complete human genome assembly. In addition, scientists are working to generate T2T genome assemblies for tomato, corn, lettuce, diploid, and triploid banana varieties.
Tools for Telomere-to-telomere Assembly
Verkko accelerates the T2T genome assembly process. Developed by a team from the Telomere-to-Telomere consortium, Verkko revolutionizes the T2T genome assembly process by automatically integrating ultra-long Oxford Nanopore sequencing reads with high-resolution assembly maps constructed from long and accurate PacBio High.
Verkko is a graph-based iterative pipeline that automates the assembly process, making it more accessible and efficient. The pipeline starts with a multiplex de Bruijn graph constructed from long and accurate reads and progressively simplifies the graph by integrating ultra-long reads and haplotype-specific markers. This approach leads to a staged diploid assembly of two haplotypes, with many chromosomes automatically assembling from telomere to telomere.
Human genome assembly HG0021 exemplifies the impact of the Verkko assembly process. Verkko assembled 20 of the 46 diploid chromosomes without gaps, achieving an impressive 99.9997% accuracy. This assembly is now a valuable resource for human genomics and has set the benchmark for T2T genome assembly projects in other organisms.
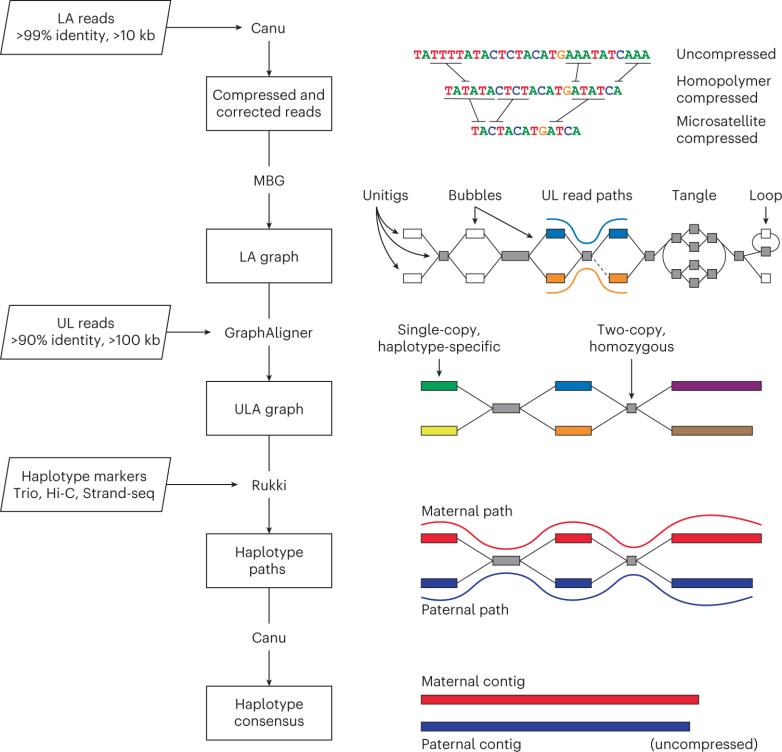
Verkko assembly workflow. (Rautiainen M et al., 2023)
Case in Point: Telomere-to-telomere Genome Assembly in Tomato and Maize
The applicability and potential of T2T genome assembly become clear when we consider recent achievements in plant genomics. The tomato (Solanum lycopersicum Heinz 1706) and maize (Zea mays B73) genomes are prime examples of how this technology is revolutionizing the field.
Telomere-to-Telomere Genome Assembly in Tomato
In the case of the tomato genome, after several manual interventions, the Verkko assembler was able to generate a complete T2T genome assembly with only one gap. This high-quality assembly was constructed from duplex data and then parsed using ultra-long simplex reads. The basic accuracy of the final assembly exceeded 99.999% (Q50), demonstrating the potential of this technology.
Telomere-to-Telomere Genome Assembly in Maize
Sequencing of the maize genome allowed the assembly of the complete T2T genome using Ultra-Long Oxford Nanopore Technology (ONT) and PacBio HiFi reads. This 2,178.6 Mb T2T Mo17 genome reveals structural features of all repeat regions in the maize genome. The assembly completely crosses every chromosome in a single overlapping cluster, marking a significant advance in understanding complex plant genomes.
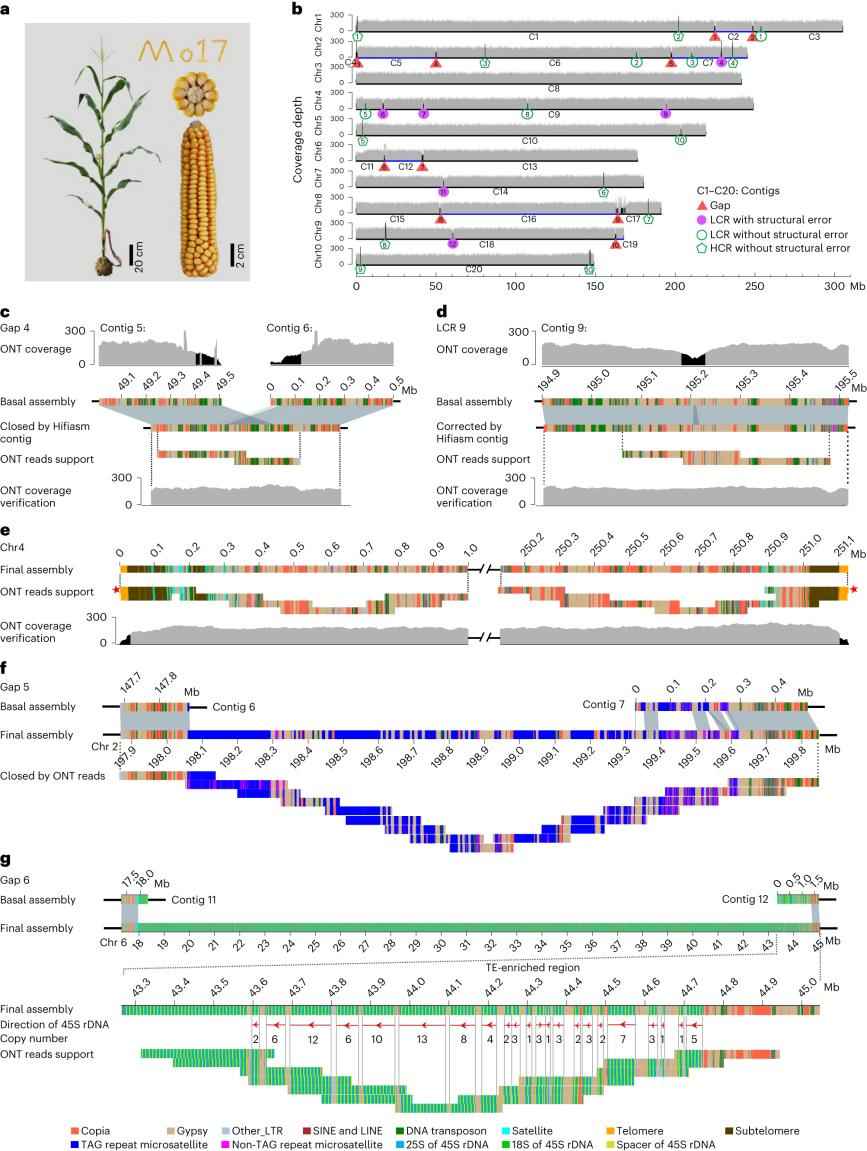
Telomere-to-telomere assembly of the Mo17 genome. (Chen J et al., 2023)
learn more:
References
- Rautiainen, Mikko, et al. "Telomere-to-telomere assembly of diploid chromosomes with Verkko." Nature Biotechnology (2023): 1-9.
- Chen, Jian, et al. "A complete telomere-to-telomere assembly of the maize genome." Nature Genetics (2023): 1-11.